RACHITT: Shuffling towards highly evolved genes
By Anthea Hammond, Ph.D.
Generation of genetic diversity through DNA recombination is critical to species adaptation and evolution. Exploiting this feature of natural evolution to molecularly breed 'improved proteins' with desired characteristics is an appealing approach for the development of novel pharmaceuticals, enzymes, DNA vaccines, modified viruses and gene therapy vehicles.
In April's edition of Nature Biotechnology, scientists from Enchira Biotechnology Corporation present a novel method for the directed evolution of proteins (1) which generates unparalleled genetic diversity; promising significant improvements to molecular breeding strategies.
The field of molecular breeding was greatly advanced in 1994 with the introduction of 'sexual PCR', a technique which generates diverse recombinant genes in vitro from a set of parental genes by 'DNA shuffling' (2). Essentially, a population of related parental genes is randomly fragmented by DNase I and allowed to hybridize, leaving overhangs which are extended by PCR techniques. Recombination occurs when a fragment derived from one template primes another with a different sequence. Repeated cycles of overlap extension yield full-length genes containing novel combinations of the parental mutations. Point mutations are introduced during the shuffling process, and variations of the technique have achieved a low and controlled mutagenesis rate.
Initially, shuffling was performed on single parental sequences; mutations for recombination being produced either by PCR mutagenesis or by the point mutagenesis associated with the shuffling process. More recently, the rich source of diversity present in naturally homologous genes has been exploited as a basis for DNA shuffling. Provided these genes share sufficient homology to cross-prime each other, they can be 'family-shuffled'. Roughly 50-fold improvements in gene function have been achieved with this method.
Until now, the sexual PCR method of DNA shuffling, and variations of it, have been the only in vitro methods shown to generate improved recombinant proteins. However, there are limitations to this approach. Most DNA shuffled proteins average just 4 crossovers per round of shuffling, limiting the potential diversity of variants produced. Furthermore, linkage effects mean that crossovers in regions of identity shorter than 15 bases are rare, resulting in lengthy tracts of sequence derived from an individual parent gene. In some cases, DNA shuffling libraries can still contain up to 100% unshuffled parental wild-type clones after the shuffling attempt.
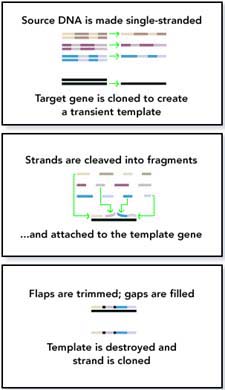 The novel method developed by the Enchira researchers sets new standards in the frequency and resolution of recombination events in family-shuffled libraries. Their technique, called 'random chimeragenesis on transient templates,' or RACHITT, also relies on homology between two related genes. Random fragments of a parental gene are hybridized with a single stranded homologue which acts as a transient scaffold template. Unhybridized flaps of parental fragments, which result from overlaps with adjacent fragments or mismatches with the scaffold, are trimmed with nucleases. By filling in the gaps between hybridized fragments, scaffold template sequences are incorporated into the nascent strand, generating a mosaic-like chimera. Finally, the transient scaffold template is destroyed and a double stranded chimeric DNA is amplified by PCR.
The novel method developed by the Enchira researchers sets new standards in the frequency and resolution of recombination events in family-shuffled libraries. Their technique, called 'random chimeragenesis on transient templates,' or RACHITT, also relies on homology between two related genes. Random fragments of a parental gene are hybridized with a single stranded homologue which acts as a transient scaffold template. Unhybridized flaps of parental fragments, which result from overlaps with adjacent fragments or mismatches with the scaffold, are trimmed with nucleases. By filling in the gaps between hybridized fragments, scaffold template sequences are incorporated into the nascent strand, generating a mosaic-like chimera. Finally, the transient scaffold template is destroyed and a double stranded chimeric DNA is amplified by PCR.
The authors use RACHITT to evolve the gene dszC, encoding dibenzothiophene mono-oxygenase (DBT-MO), a key enzyme of the diesel biodesulfurization pathway used in refinery-level biodesulfurization of fossil fuels. They successfully recombine dszC genes from two different bacteria, one with a higher substrate affinity and/or substrate range, and the other with a higher specific reaction rate for a particular substrate type. The two genes are 89.9% identical, encoding proteins with 38 amino acid differences. After screening relatively few clones (175 of the 35,000 clone library) without selection, chimeras with 60-320% improvement in flux through the enzyme pathway were isolated. When applying a growth selection by screening the library on a low-sulfur diesel substrate, variants simultaneously improved in both rate and extent of substrate range were isolated at relatively high frequency. Furthermore, the authors were able to evolve substrate specificity of the progeny enzymes, breeding variants with 20-fold better conversion of a non-natural substrate.
At the molecular level too, RACHITT has significant advantages over other methods of DNA shuffling. Analysis of 22 unselected genes revealed a predominance of very highly mosaic clones. The average number of crossovers was 14, several-fold greater than that observed with sexual PCR-based DNA shuffling. And on average, RACHITT-derived genes contained more than 4 recombination events between alleles closer than 10 bases apart. This unprecedented frequency is a significant advance towards generating libraries which maximally exploit the genetic diversity inherent in the parental genes.
RACHITT produced relatively few inactive proteins, no unshuffled parental clones, and no siblings (duplicate occurrences of the same chimeric gene). A fundamental concern for DNA shuffling approaches is that while strategies which create a more diverse library increase the potential for identifying improved genes, they also create more progeny with phenotypes less fit than the parent, necessitating a highly efficient screening strategy. But RACHITT shuffles DNA more completely and produces fewer inactive proteins than previous methods, enabling more diverse libraries to be handled. Indeed the authors have also produced shuffled libraries and evolved clones from 4-14 parents simultaneously.
The impressive features of RACHITT represent advances in critical determinants of the evolutionary potential of DNA-shuffled libraries. RACHITT, then, may allow scientists to more effectively examine increasingly diverse permutations of mutations; breeding new generations of improved proteins.
References:
- Coco WM, Levinson WE, Crist MJ, Hektor HJ, Darzins A, Pienkos PT, Squires CH, Monticello DJ. DNA shuffling method for generating highly recombined genes and evolved enzymes. Nat. Biotech. 19: 354-359 (2001).
- Stemmer WP. Rapid evolution of a protein in vitro by DNA shuffling. Nature 370: 389-391 (1994).
About the author: Anthea Hammond, Ph.D., is a research scientist interested in virology and infectious disease. She currently works in the Molecular Medicine Program at the Mayo Clinic.
Sign up for our free newsletter.
Send comments to Bioresearch Online's content manager at contentmanager@bioresearchonline.com.
